Ph.D., Russian Cancer Research Center, 1979
how cells move: molecular hardware of cell motility
Individual cells in multicellular organisms are able to move. This ability is essential for virtually every aspect of the whole body functioning. Cell motility is a complicated multi-step process, which begins from making a decision to move, which occurs at the cell surface. When surface receptors receive external signals, they initiate a signaling cascade inside the cell and issue commands to the cytoskeleton to generate motility. Cytoskeleton is a complex of detergent-resistant cytoplasmic components, which may be considered as molecular hardware for motility. The major question of our research is how this molecular hardware works.

Fig. 1: Main components of the fibroblast cytoskeleton.
Actin filaments (yellow), microtubules (red), and
intermediate filaments (blue).
Platinum replica\electron microscopy.
The design of an unknown machine can be understood based on how its elements are structurally arranged, how they move during action, and what happens if some element is missing. Our main experimental approach is based on the same idea applied to cells and molecules. We use platinum replica electron microscopy (EM) to analyze the structural organization of the cytoskeleton at the nanometer scale level (Fig. 1). This EM technique reveals a fascinating world of the cell interior and frequently gives simple answers to complicated questions. Since EM is not applicable to living cells, to see the machinery in action we use correlative analysis, in which the dynamic observation of a cell is followed by EM of the same cell (Fig. 2).
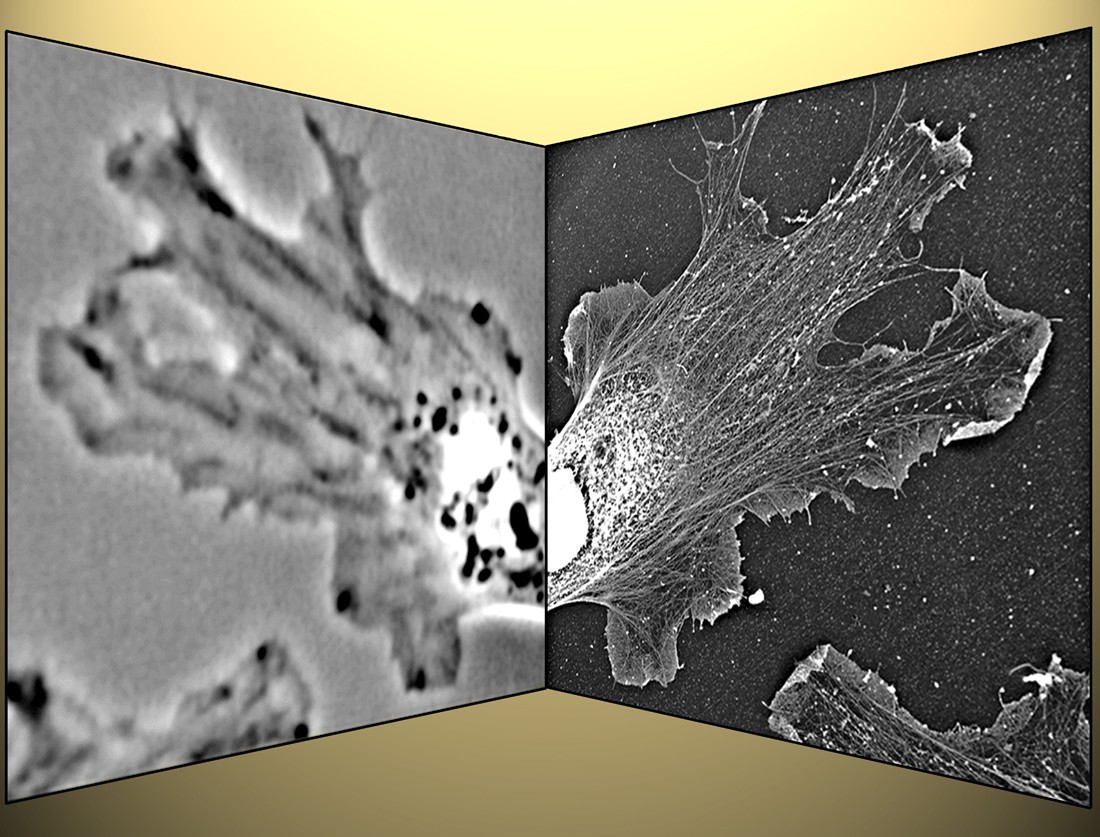
Fig. 2: Correlative light and electron microscopy.
After phase-contrast microscopy of a living Xenopus fibroblast (left),
the sample was processed for platinum replica electron microscopy,
and the cell is re-localized (right). This technique allows to correlate
motility with the cytoskeleton organization.
This approach allows establishing functional connections between cytoskeletal dynamics and supramolecular organization. Functional perturbations of specific molecules give further insight into details of the molecular design of cellular motile machinery.
Cell locomotion consists of repeated cycles of leading edge protrusion followed by cell body translocation. The primary focus of our research is to understand the mechanisms of leading edge protrusion. Protrusion is driven by polymerization of actin, the major structural and functional element of the cytoskeleton. Actin filaments through interaction with different sets of accessory proteins are able to form a remarkable variety of superstructures with different design and functions. Lamellipodia and filopodia are the two major protrusive organelles with strikingly different structural organization and different sets of molecular players. Different cell types use these two organelles to a different extent. Lamellipodia, which are broad, flat protrusions are filled with a branched network of actin filaments (Fig. 3).
Fig. 3: Dendritic actin network in fibroblast lamellipodium.
Individual families of branching filaments are highlighted
in different colors.
The current model of actin dynamics in lamellipodia describes it as a cycle of dendritic nucleation, elongation, capping, and depolymerization of actin filaments. Filopodia, which are thin cellular processes, contain a tight bundle of parallel actin filaments elongating at the tip and depolymerizing from the rear (Fig. 4).
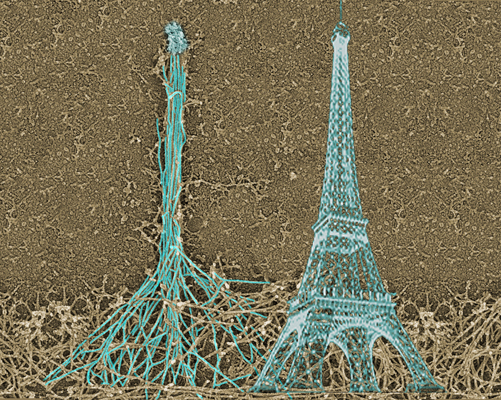
Fig 4: Assembly of the filopodial bundle occurs at the tip,
similar to how the tower is built. As a result, the history
of the construction is imprinted in the design of the
structure allowing to understand how the filopodium was built.
Although basic models for the leading edge protrusion have been formulated, many questions remain about the molecular mechanisms of this process and specific roles of individual molecules. It is also unclear how design of the protrusive machinery varies depending on the cell type and on physiological or pathological conditions. Our strategy is to employ a combination of powerful structural, dynamic, and functional approaches to address these questions. When these pieces of information come together, they always produce something novel and exciting.
BIOL 2010 - Cell biology
BIOL 4024 - Cell motility and cytoskeleton
Svitkina, T.M. 2023. Filopodia and lamellipodia. In Encyclopedia of Cell Biology (Second Edition). Vol. 3. R.A. Bradshaw, G.W. Hart, and P.D. Stahl, editors. Academic Press, Oxford. 197-210.
Yang, C., P. Colosi, S. Hugelier, D. Zabezhinsky, M. Lakadamyali, and T. Svitkina. 2022. Actin polymerization promotes invagination of flat clathrin-coated lattices in mammalian cells by pushing at lattice edges. Nat Commun. 13:6127.
Svitkina, T. 2022. Imaging cytoskeleton components by electron microscopy. Methods Mol Biol. 2364:25-52.
Svitkina, T.M. 2021. Cytoskeleton | Actin organization☆. In Encyclopedia of Biological Chemistry III (Third Edition). Vol. 5. J. Jez, editor. Elsevier, Oxford. 154-166.
Efimova, N., C. Yang, J.X. Chia, N. Li, C.J. Lengner, K.L. Neufeld, and T.M. Svitkina. 2020. Branched actin networks are assembled on microtubules by adenomatous polyposis coli for targeted membrane protrusion. J Cell Biol. 219: e202003091.
Svitkina, T.M. 2020. Actin cell cortex: Structure and molecular organization. Trends Cell Biol. 30:556-565.
Yang, C., and T.M. Svitkina. 2019. Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol. 21:603-613.
Chikina, A.S., T.M. Svitkina, and A.Y. Alexandrova. 2019. Time-resolved ultrastructure of the cortical actin cytoskeleton in dynamic membrane blebs. J Cell Biol. 218:445-454.
Svitkina, T.M. 2018. Ultrastructure of the actin cytoskeleton. Curr Opin Cell Biol. 54:1-8.
Svitkina, T. 2018. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol. 10:a018267.
Shutova, M.S., and T.M. Svitkina. 2018. Mammalian nonmuscle myosin II comes in three flavors. Biochem Biophys Res Commun. 506:394-402.
Efimova, N., and T.M. Svitkina. 2018. Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J Cell Biol: 217:1827-1845.
Shutova, M.S., S.B. Asokan, S. Talwar, R.K. Assoian, J.E. Bear, and T.M. Svitkina. 2017. Self-sorting of nonmuscle myosins IIA and IIB polarizes the cytoskeleton and modulates cell motility. J Cell Biol. 216:2877-2889.
Svitkina, T.M. 2017. Platinum replica electron microscopy: Imaging the cytoskeleton globally and locally. Int J Biochem Cell Biol. 86:37-41.
Efimova, N., F. Korobova, M.C. Stankewich, A.H. Moberly, D.B. Stolz, J. Wang, A. Kashina, M. Ma, and T. Svitkina. 2017. betaIII spectrin is necessary for formation of the constricted neck of dendritic spines and regulation of synaptic activity in neurons. J Neurosci. 37:6442-6459.
Chia, J.X., N. Efimova, and T.M. Svitkina. 2016. Neurite outgrowth is driven by actin polymerization even in the presence of actin polymerization inhibitors. Mol Biol Cell. 27:3695-3704.
Shutova, M.S., W.A. Spessott, C.G. Giraudo, and T. Svitkina. 2014. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 24:1958-1968.
Ong, K., C. Wloka, S. Okada, T. Svitkina, and E. Bi. 2014. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun. 5:5698.
Jones, S.L., F. Korobova, and T. Svitkina. 2014. Axon initial segment cytoskeleton comprises a multiprotein submembranous coat containing sparse actin filaments. J Cell Biol. 205:67-81.
Shutova, M., C. Yang, J.M. Vasiliev, and T. Svitkina. 2012. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One. 7:e40814.
Hoelzle, M.K., and T. Svitkina. 2012. The cytoskeletal mechanisms of cell-cell junction formation in endothelial cells. Mol Biol Cell. 23:310-323.
Yang, C., and T. Svitkina. 2011. Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol. 13:1012-1013.
Yang, C., and T. Svitkina. 2011. Filopodia initiation: Focus on the Arp2/3 complex and formins. Cell Adh Migr. 5:402-408.
Gorelik, R., C. Yang, V. Kameswaran, R. Dominguez, and T. Svitkina. 2011. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell. 22:189-201.
Collins, A., A. Warrington, K.A. Taylor, and T. Svitkina. 2011. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 21:1167-1175.
Korobova, F., and T. Svitkina. 2010. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 21:165-176.
Yang, C., M. Hoelzle, A. Disanza, G. Scita, and T. Svitkina. (2009) Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One, 4:e5678.
Korobova, F. and T. Svitkina. (2008) Arp2/3 Complex is important for filopodia formation, growth cone motility and neuritogenesis in neuronal cells. Mol. Biol. Cell, 19: 1561-1574.
Yang, C., L. Czech, S. Gerboth, S. Kojima, G. Scita, and T. Svitkina. (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biology 5:e317.
American Society for Cell Biology
American Association for the Advancement of Science

